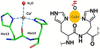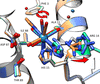issue contents
May 2024 issue
scientific commentaries
MATERIALS | COMPUTATION
Crystal structues exhibiting disorder still present a barrier for many computational methods. Dittrich et al. [(2024). IUCrJ, 11, 347–358] showcase a unified approach, tackling solid solutions, near symmetry and more.
ELECTRON CRYSTALLOGRAPHY
Over 30 years ago, it was shown that bonding between atoms has a noticeable effect on convergent beam electron diffraction patterns. The paper by Olech et al. [(2024). IUCrJ, 11, 309–324] demonstrates that its influence is also clearly present in 3D electron diffraction data, opening up new possibilities for quantum crystallography.
topical reviews
BIOLOGY | MEDICINE
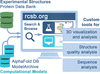
The RCSB PDB research-focused web portal at https://www.rcsb.org/ provides important tools and resources to search, visualize and analyze experimentally determined 3D biostructures alongside computed structure models of proteins predicted using artificial intelligence/machine-learning based tools.
BIOLOGY | MEDICINE
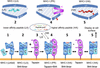
This topic review summarizes structures of chaperones complexed with MHC-I, the structural principles that govern peptide exchange and the mechanism in antigen presentation.
research papers
BIOLOGY | MEDICINE
A cytotoxin encoded by an `orphan' genetic locus in the insect pathogen Yersinia entomophaga is shown to be a functional part of an ABC toxin complex, and the structures of both the RHS-repeat-containing `shell' and the free toxin alone are determined by X-ray crystallography. The structure of the toxin indicates that it most likely functions by directly modifying actin in the target cell.
ELECTRON CRYSTALLOGRAPHY
Download citation


Download citation


Transferable aspherical atom model dynamical refinement on precession electron diffraction data for 1-methyluracil crystals offers superior performance compared with the independent atom model and reveals that the quality of 3D electron diffraction data and dynamical refinement is already sufficient to detect minute variations of the electrostatic potential caused by bonding and intermolecular interactions.
NEUTRON | SYNCHROTRON
An X-ray absorption spectroscopy electrochemical cell was used to collect high-quality X-ray absorption spectroscopy measurements of N-truncated Cu:amyloid-β (Cu:Aβ) samples under near-physiological conditions. The geometry of binding sites for the copper binding in Aβ4–8/12/16 and the ability of these peptides to perform redox cycles in a manner that might produce toxicity in human brains were determined.
MATERIALS | COMPUTATION
Solid-state phenomena like disorder, polymorphism but also the occurrence of high-Z′ crystal structures can be linked via energy differences in `archetype crystal structures', which will permit better prediction of their occurrence.
BIOLOGY | MEDICINE
A detailed analysis of rhenium(I) organometallic covalent binding to a model protein is conducted at seven time points over 38 weeks. Changes in the protein structure induced at the Re binding sites over the time series as well as the relationship between the proximity of the solvent channels to the residues containing the highest-occupied Re are described.
PDB reference: HEWL-[Re(CO)3-Im], 8qcu
BIOLOGY | MEDICINE
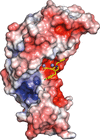
A structural analysis of several metal-ion binders that inhibit viral endonucleases is performed.
BIOLOGY | MEDICINE
The first crystal structure of phytoplasma immunodominant membrane protein is reported and structural analysis revealed its potential for actin binding.
PDB reference: phytoplasma immunodominant membrane protein, 8j8y
BIOLOGY | MEDICINE
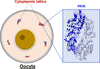
The human peptidylarginine deiminase type VI (PAD6) is essential in oocyte and embryonic development as a component of the supramolecular assemblies called cytoplasmic lattices. The crystal structure presented here suggests PAD6 assembles as a dimer resembling other PADs, albeit with compromised abilities to bind Ca2+ and substrates. This aligns with existing in vitro data which indicate an enzymatically inactive isoform of PAD.
PDB reference: Human PAD6 phosphomimic mutant V10E/S446E, apo, 8ql0
PHYSICS | FELS
A kinetics-informed neural-network method (KINNTREX) is designed to analyze a time series of difference maps from a time-resolved X-ray crystallographic experiment.
NEUTRON | SYNCHROTRON
Total X-ray scattering measurements of dilute solutions are uniquely well suited to address the structural basis for the effects of solvation, exemplified by the success of small-/wide-angle X-ray scattering studies of macromolecules in solution. Herein, we extend this methodology to the high-energy X-ray regime by developing a theoretical framework to describe the differential solution scattering experiment, supported by numerical simulation and experiment.


 journal menu
journal menu




 access
access